 |
| translation of messenger RNA |
Outline
In outline, the process of protein synthesis is as follows. Amino acids are activated for protein synthesis by amino acid synthetases which attach the amino acids to their cognate tRNA molecules. The smaller ribosomal subunit and then the larger ribosomal subunit attach to messenger RNA at the 5’ end or near the initiating codon. Translation then begins at an initiation codon with the assistance of initiation factors. During the process of protein synthesis, the activated amino acids to be incorporated into the peptide chain are specified by threebase codon-anticodon pairings between the messenger and aminoacyltRNA. Elongation of the peptide chain terminates on recognition of one of the three termination codons, the ribosomes and messenger dissociate, and the newly synthesized peptide is released.
Now we will discuss about translation in detail.
Various events of protein synthesis can be studied under the following headings :
A. Aminoacylation of tRNA (Formation of Aminoacyl-tRNA)Amino acids alone do not come to the ribosome to be incorporated into protein. Instead, they are brought to the ribosome by their appropriate tRNA. The first step in incorporating an amino acid into a protein involves the amino acids attachment to its correct tRNA. This involves the following two steps:
(a) Activation of amino acids. Each of the 20 amino acids occur in the cytoplasm in an inactive state. Each amino acid before its attachment with its specific tRNA is activated by a specific activating enzyme known as the aminoacyl synthetase and ATP. The free amino acids react with ATP, resulting in the production of aminoacyl adenylate and pyrophosphate :AA + ATP + Enzyme AA ~ AMP—Enzyme + PP Amino Aminoacyl Aminoacyl adenylate Pyrophosphate acid synthetase enzyme complex The reaction product aminoacyl adenylate (or aminoacyl adenosine monophosphate) is bound to the enzyme. The cell has at least 20 aminoacyl synthetase enzymes for the 20 amino acids. Each enzyme is specific and it attaches with the specific amino acid without any error.
 |
| tRNA activation |
(b) Attachment of activated amino acid to tRNA. The aminoacyl adenylate remains bounded with enzyme until it collides with the specific tRNA molecule and its synthetase is recognized by dihydrouridine (DHU) loop of specific tRNA. Then, amino acid residue of aminoacyl adenylate is transferred to amino acid attachment site of tRNA where its carboxyl group forms bond or linkage with the 3-OH group of the ribose of the terminal adenosine at CCA end of tRNA. As a result AMP and enzyme are released and a final product aminoacyl tRNA is formed by the following method : AA—AMP—Enzyme + tRNA A A — tR N A + A M P + Enzyme Aminoacyl adenylate and enzyme Amioacyl—tRNA The aminoacyl-tRNA moves towards the site of protein synthesis.
Further, recent evidence suggests that the aminoacyl synthetase enzyme checks for correct binding before the release of the charged tRNA (i.e., tRNA-AA). If an error has been made, the wrong amino acid is removed and the correct one is attached.
 | ||
| Activated tRNA |
B. Stages of Polypeptide Synthesis
Polypeptide synthesis in prokaryotes and eukaryotes follow the same overall mechanism with few exception which we will discuss in detail later
An important feature of initiation of polypeptide synthesis is the use of specific initiating tRNA molecule. In prokaryotes this tRNA molecule is acylated with the modified amino acid N-formyl methionine (f Met);this acylated tRNA recognizes the codon AUG and is used for initiation.The tRNAf Met molecule is first acylated (or charged) with methionine and an enzyme (found only in prokaryotes) adds a formyl group to the amino group of the methionine.
In eukaryotes, the initiating tRNA molecule is charged with methionine. The use of these initiator tRNA molecules means that while being synthesized, all
prokaryotic proteins have N-formylmethionine at the amino terminus and all eukaryotic proteins have methionine at the amino terminus. However, these amino acids are frequently altered or removed later by the activity of a hydrolytic enzyme (this is called processing).Since in protein synthesis the peptide chain always grows in a sequence from the free terminal amino (–NH2) group towards the carboxyl (–COOH) end, so the function of methionine-tRNA is to ensure that protein are synthesized in that direction and thus the terminal amino group is blocked leaving only the —COOH group available to react with the —NH2 group of the second amino acid (AA2). In this way the synthesis of protein chain follows in the correct sequence.
In eukaryotes the initiation of the peptide chain starts with some initiation factors and at least ten initiation factors have been identified in red blood cells (= reticulocytes). They are named by putting a prefix 'e' to signify their eukaryotic origin. These factors are eIF1, eIF2, eIF3, eIF4A, eIF4B, eIF4C, eIF4D, eIF5 and eIF6.
Now, let us discuss, in brief, the polypeptide synthesis in eukaryotes :
Initiation: This process involves the following steps : (i) GTP binds to eIF2 and to this complex becomes associated Met-tRNA to form a ternary complex, i.e., Met-tRNA-eIF2-GTP. The protein eIF2 in mammals and wheat plant has three subunits, namely α, β and γ; eIF2 α binds to GTP; eIF2γ binds to Met-tRNA and eIF2β may be a recycling factor. (ii) The ternary (=of three components) complex associates with 40S subunit to form 43 initiation complex. (iii) The mRNA at its 5' end binds with 43 initiation complex. This reaction depends on eIF3 and the binding of mRNA is assisted by eIF4F, eIF4A, eIF4B and a high energy bond of ATP. (iv) After association of the 5' end of mRNA, initiation complex moves towards 3' end in search of initiation codon AUG, and then also associates with 60S subunit. Association of 60S subunit with the initiation complex requires the factor eIF5, because it helps in releasing eIF2 and eIF3 (60S subunit cannot join, if these two initiation factors are not released). The eIF2 is released as a binary complex, eIF2-GDP. The 40S-60S joining reaction really depends on eIF4C. The GTP of the initiation complex is hydrolyzed, when 60S subunit joins
 |
| Initiation |
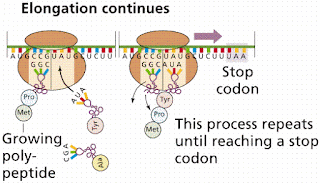
2. Elongation. For the process of elongation of the peptide chain the next (second) codon in mRNA is base paired by the appropriate AA-tRNA at the A site. This requires the presence of EF1 (elongation factor1) and energy (i.e., one GTP is hydrolyzed to GDP). A peptide is then formed to join the amino group of aa-tRNA with the carboxyl group of p-tRNA. This is achieved by peptidyl transferase enzyme present in the 60S ribosomal subunit. The discharged tRNA in the P site is immediately ejected out. There upon EF2 (also called translocase or G factor) causes the translocation of newly formed P-tRNA and its codon from A to P site; again one GTP is hydrolyzed to GDP to provide the necessary energy. In this way the polypeptide chain continues to be elongated.
3. Termination. The termination of growing polypeptide chain occurs when a termination codon comes to occupy the 'A' site. There is no tRNA for these codons. The releasing factor 1 or 2 with the help of peptidyl transferase releases the polypeptide from the P-tRNA. This also causes the release of tRNA from the 'P' site and dissociation of 80S ribosome into 60S and 40S subunits. In eukaryotic systems, only one release factor is known, i.e., eRFI. GTP seems to be necessary for binding of this factor to ribosome. GTP is cleaved after termination step has occurred which may be essential for the release of eRFI from the ribosome.
 |
| termination |






No comments:
Post a Comment
Thank you for commenting.