Learning objects:
DNA ligase are used to join the fragments on lagging strand and thus they fill the gaps.
Primers
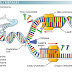
DNA polymerase can not initiate the process of replication it self.They are required RNA primer.So before actual RNA replication starts, short RNA oligonucleotide segments,called RNA primers have to be synthesized by DNA primase enzyme utilizing ribo_nucleoside triphosphate.This RNA primer has a particular sequence of bases as one of the template strand.The primers are about 10 nucleotides long in eukaryotes and they are made at intervals on lagging strand which are later replaced by deoxyribonucleotides to form okazaki fragments.
In bacteria two different enzymes are known to synthesize primer RNA.These are RNA polymerase(on the leading strand) and DNA primase (on the lagging strand)
Proteins involved in opening of DNA helix
The following three types of proteins are needed to help the DNA double helix to open and to provide exposed DNA template for the DNA polymerase to copy.
DNA helicases
DNA helicases are ATP dependent unwinding enzymes which promote separation of the two parental strands and thus form a replication fork.DNA helicase hydrolyze ATP when they are bound to single strand of DNA
Helix_destabilizing strand or single strand DNA_binding proteins(SSBPs)
These proteins prevent the opened DNA helix from rewinding.SSB proteins bind to exposed DNA strands without covering the bases, which, therefore remain available for the templating process.
Topoisomerases (DNA gyrases)
The activity of helicase causes the supercoiling of DNA duplex ahead of the replication fork.Enzymes called topoisomerases relax the supercoil by attaching and then nicking one of the strands and rotating it through the unbroken strand.Topoisomerases have two types :topoisomerase 1 and topoisomerase 2
Replisome N.K. Sinha and A.K.ornberg have suggested that the DNA polymerases, RNA primases and helicases may be associated with one another to form a multienzyme complex — the replisome that carries out the synthesis of leading and lagging strands in a coordinated fashion. Such a complex would be highly processive and assure rapid replication of the DNA
Note: in our next post we will discuss about the mechanism of replication in prokaryotes and eukaryotes.
- nuclease enzymes and their functions
- polymerases, its types both in pro and eukaryotes
- role of ligases
- role of helicase enzyme
- role of SSSB
- role of topoisomerase
- Replisome
Nuclease enzymes
The nuclease enzymes act to hydrolyze or break down a polynucleotide chain into its component nucleotides.A polynucleotide is held together by3',5' phosphodiester bonds and a nuclease enzyme will attack either the 3'or 5' end of this linkage.The nuclease enzymes may be of the following two kinds
- Exonuclease enzyme
A nuclease enzyme which begins its attack from a tree end or terminal points of a polynucleotide is called exonuclease.
- Endonuclease enzyme
Polymerase or replicase enzymes
A polymerase enzyme catalyses the formation of a polymer.Polymerase enzymes are of different types.In prokaryotes these are DNA polymeraseI,DNA polymerase II, DNA polymerase III.Polymerase I and II are meant for DNA repair and polymeraseIII is meant for actual DNA replication.
 |
| DNA polymerase |
DNA polymerase I
This enzyme was isolated round 1960 by Arthur Kornberg and was the first enzyme suggested to be involved in DNA replication.It is also called Kornberg enzyme.DNA polymerase I is mainly involved in removing RNA primers.DNA polymerase I acts as exonuclease and also performs the role of proof reading or editing function.
DNA polymerase II
This enzyme resembles DNA polymerase 1 in its activity, but it is a DNA repair enzyme.
DNA polymerase III
It is the most important enzyme of replication.It is a holo_enzyme having ten sub_units such as alpha ((), beta (β) , psilon (ε), theta (θ), tau (τ) gamma (γ), delta (δ), delta dash (δ´), chi (χ), and psi (ψ) .All these sub_units have different functions.The core enzyme comprises three sub_units alpha, beta and theta.Remaining 7 sun_units increase processivity.
Eukaryotic DNA polymeraseEukaryotic cells have just DNA polymerase 3 this enzyme has further 5 types.These are
- alpha(α) DNA polymerase is also known as cytoplasmic polymerase.Itis found both in nucleus and cytoplasm.
- DNA polymerase beta(β): This is also called nuclear polymerase and found only i vertebrates.
- DNA polymerase gamma:This is called mitochondrial polymerase and is encoded in the nucleus
- DNA polymerase delta:This enzyme is found in mammalian cells and is PCNA dependent for DNA synthesis processivity.
- DNA polymerase epsilon: It is also known as PCNA dependent enzyme and found in mammalian HeLa cells.
DNA ligase are used to join the fragments on lagging strand and thus they fill the gaps.
Primers
 |
| RNA primer |
In bacteria two different enzymes are known to synthesize primer RNA.These are RNA polymerase(on the leading strand) and DNA primase (on the lagging strand)
Proteins involved in opening of DNA helix
The following three types of proteins are needed to help the DNA double helix to open and to provide exposed DNA template for the DNA polymerase to copy.
DNA helicases
DNA helicases are ATP dependent unwinding enzymes which promote separation of the two parental strands and thus form a replication fork.DNA helicase hydrolyze ATP when they are bound to single strand of DNA
Helix_destabilizing strand or single strand DNA_binding proteins(SSBPs)
 |
| SSSB |
These proteins prevent the opened DNA helix from rewinding.SSB proteins bind to exposed DNA strands without covering the bases, which, therefore remain available for the templating process.
Topoisomerases (DNA gyrases)
The activity of helicase causes the supercoiling of DNA duplex ahead of the replication fork.Enzymes called topoisomerases relax the supercoil by attaching and then nicking one of the strands and rotating it through the unbroken strand.Topoisomerases have two types :topoisomerase 1 and topoisomerase 2
Replisome N.K. Sinha and A.K.ornberg have suggested that the DNA polymerases, RNA primases and helicases may be associated with one another to form a multienzyme complex — the replisome that carries out the synthesis of leading and lagging strands in a coordinated fashion. Such a complex would be highly processive and assure rapid replication of the DNA
Note: in our next post we will discuss about the mechanism of replication in prokaryotes and eukaryotes.






No comments:
Post a Comment
Thank you for commenting.