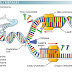
Recombinant DNA Technology (rDNA Tech) or genetic engineering is concerned with the manipulation of genetic materials towards desired end in a directed way. It is also known as gene cloning.In this process the DNA molecules are isolated and cut into pieces by one or more specialised enzymes and the fragments are joined together in a desired combination and restored to a cell for replication and reproduction. Recombinant DNA, thus is a composite DNA molecule that results from the physical combination of DNA segments derived from different sources.
In other words, the Genetic engineering is biochemical manipulation of genes by which foreign genes or the genes of donor organisms are inserted into the DNA or (touted recipient organism through the plasmids of bacteria or through bacteriophages.
Historical Account of Genetic Engineering:In nature the transfer of genetic material occurs through conjugation, transformation and transduction.
Recombination is brought about by the following two processes:
(i) Natural recombination process, and
(ii) Artificial recombination process.
The natural recombination occurs during meiosis when similar nucleotide sequences in DNAs of homologous chromosomes come closer, and exchange segments and then rejoin. Thus a new arrangement of genes results from this recombination.
The artificial recombination process involves unrelated organisms. The genetic material of one organism is inserted into a specialized DNA molecule of another organism giving rise to recombinant or rDNA.
The history of modern genetic engineering goes back to mid 1970s when it became possible to cut DNA into pieces and transfer a particular piece of DNA containing specific information from donor organism to the DNA of the recipient organism.
And year-wise landmarks of the genetic engineering are given here as follows:
1970 Smith and Nathans discovered a new class of enzymes, the restriction enzymes that act as chemical scissors and cut a DNA molecule into smaller fragments.1972 Berg and others combined DNAs from two viruses to produce what is called recombinant DNA (rDNA).
1973 Cohen and Boyer inserted recombinant DNAs into host bacteria that reproduced or cloned foreign DNAs. With this, the age of genetic engineering begins.
1977 Genetech, one of the first genetic engineering companies starts the biosynthesis of important drugs by rDNA Technology.
1977 Sanger and Gilbert independently discovered techniques for rapid sequencing of nucleotides in DNA molecules.
1982 Human insulin produced by rDNA is marketted under the trade name ‘Humulin’.
1983 Tracy Moreno, a ten year old girl born with growth hormone deficiency, grows 5 inches in one year of treatment with engineered hormones. Human growth hormone genes are inserted into mouse embryo producing a giant mouse. For the first time a human gene functions in another animal.
1997 Scientists remove the DNA containing nucleus from a female’s egg and replace it with a nucleus from a different animal of the same species. The scientists then place that egg into the uterus of a third animal.
The result, first demonstrated by the birth of a cloned sheep named Dolly, is the birth of an animal that is nearly genetically identical to the animal from which the nucleus was obtained. Such an animal is genetically unrelated to the surrogate mother.
Such experiments were also done to produce the clone of human beings but....
In February, 2003 the Human Cloning Prohibition Act was passed which made it a crime for any one, public or private, to conduct somatic cell nuclear transplantation on fertilized or unfertilized human egg cell for reproductive or therapeutic purposes. The penalty for engaging in such research is US $ 1 million fine and a jail term for 10 years.
Steps of Genetic Engineering: The gene cloning or recombinant DNA formation requires the following
1. DNA fragments to be cloned or target gene sequence of desired type.
2. Restriction endonucleases for cutting DNA molecule into fragments.
3. Cloning vector.
4. DNA ligase enzyme for splicing DNA segments.
5.Expression system
The major steps involved in the genetic engineering are as follows
1. To break open the living cells:Several methods are available to break open the living cells. One of the popular methods involves mechanical shearing, the cells in a blender and then treating them with a detergents.
2. Isolation and identification of desired genes or DNA sequence:
The genetic information is stored in DNA. Since the DNA molecules are much longer than all other molecules found in the cells, it has become possible to develop technique of purifying DNA. DNA molecules are spooled on a glass rod. The glass rod bearing the DNA molecules is then lifted out from the mixture of broken cells.
3. Cutting of the DNA molecules into segments containing specific genes from the rest of DNA:
DNA is cut into gene size segments with the help of molecular scissors,called restriction endonucleases.
The restriction enzymes, which cut DNA at defined sites, represent one of the most important groups of enzymes for the manipulation of DNA. These enzymes are found in bacterial cells, where they function as part of a protective mechanism called the restriction-modification system. In this system the restriction enzyme hydrolyses any exogenous DNA that appears in the cell. To prevent the enzyme acting on the host cell DNA, the modification enzyme of the system (a methylase) modifies the host DNA by methylation of particular bases in the restriction enzyme’s recognition sequence, which prevents the enzyme from cutting the DNA.
Restriction enzymes are of three types (I, II, or III). Most of the enzymes commonly used today are type II enzymes.These enzymes are nucleases, and as they cut at an internal position in a DNA strand (as opposed to beginning degradation at one end) they are known as endonucleases. Thus, the correct designation of such enzymes is that they are type II restriction endonucleases, although they are often simply called restriction enzymes. In essence they may be thought of as molecular scissors.
Restriction enzyme nomenclature
Restriction enzyme nomenclature is based on a number of conventions. The generic and specific names of the organism in which the enzyme is found are used to provide the first part of the designation, which comprises the first letter of the generic name and the first two letters of the specific name.Thus, an enzyme from a strain of Escherichia coli is termed Eco, one from Bacillus amyloliquefaciens is Bam, and so on.
Use of restriction endonucleases
Restriction enzymes are very simple to use -- an appropriate amount of enzyme is added to the target DNA in a buffer solution, and the reaction is incubated at the optimum temperature (usually 37◦C) for a suitable length of time. Enzyme activity is expressed in units, with one unit being the amount of enzyme that will cleave one microgram of DNA in one hour at 37◦C.
Type of restriction endonucleases on the base of activityThere are two groups of restriction enzymes. Some enzymes recognise a specific nucleotide pair sequence and then cut the DNA at random i.e., at non-specific site away from the recognition site.
The other group of enzymes cleave the DNA at specific site. All restriction enzymes are sequence specific and thus the number of cuts they make in a DNA molecule depends upon the number of times, the specific sequences are repeated in DNA molecules. Different endonucleases found in different organisms recognise different nucleotide sequences and they cut DNA at different cleavage sites.
Due to the activities of restriction enzymes following types of DNA fragments are produced:
(1) blunt ends (sometimes known as flush-ended fragments), (2) fragments with protruding 3 ends, and (3) fragments with protruding 5 ends.
Enzymes such as EcoRI generate DNA fragments with cohesive or ‘sticky’ ends, as the protruding sequences can base pair with complementary sequences generated by the same enzyme. Thus, by cutting two different DNA samples with the same enzyme and mixing the fragments together, recombinant DNA can be produced. This is one of the most useful applications of restriction enzymes and is a vital part of many manipulations in genetic engineering.
3.Use of a vector
The biology of gene cloning is concerned with the selection and use of a suitable carrier molecule or vector, and a living system or host in which the vector can be propagated.
The means by which recombinant DNA is introduced into a host cell is called vector.
There are certain features that vectors must posess. Ideally they should be fairly small DNA molecules, to facilitate isolation and handling. There must be an origin of replication, so that their DNA can be copied and thus maintained in the cell population as the host organism grows and divides. It is desirable to have some sort of selectable marker that will enable the vector to be detected, and the vector must also have at least one unique restriction endonuclease recognition site to enable DNA to be inserted during the production of recombinants. Plasmids have these features and are extensively used as vectors in cloning experiment.
Plasmids:Many types of plasmids are found in nature, in bacteria and some yeasts. They are circular extrachromosomal DNA molecules carrying genes for antibiotic resistance and fertility in bacteria.
Plasmids can be classified into two groups, conjugative and non-conjugative plasmids. Conjugative plasmids can mediate their own transfer between bacteria by the process of conjugation,Non-conjugative plasmids are not selftransmissible but may be mobilised by a conjugation-proficient plasmid if their mobilising region is functional.
DNA of bacterial viruses
The DNA of bacterial virus can also be used as a vector.For example lambda phage.Lambda phage attaches to a host bacterium.The recombinant DNA is released from the virus and enters into the bacterium.The recombinant DNA replicates and many copies of the viruses are formed.Each virus in bacteriophage clone contains a copy of gene of choice.
The bacteriophage-based vectors are more specialised than plasmid vectors.Two types of bacteriophage (lambda and M13) have been extensively developed for cloning purposes
DNA ligase – joining DNA molecules
DNA ligase is an important cellular enzyme, as its function is to repair broken phosphodiester bonds that may occur at random or
as a consequence of DNA replication or recombination. In genetic engineering it is used to seal discontinuities in the sugar--phosphate chains that arise when recombinant DNA is made by joining DNA molecules from different sources. It can therefore be thought of as
molecular glue, which is used to stick pieces of DNA together. This function is crucial to the success of many experiments, and DNA ligase is therefore a key enzyme in genetic engineering.
The enzyme used most often in experiments is T4 DNA ligase, which is purified from E. coli cells infected with bacteriophage T4.
Expression system or Host An ideal host cell should be easy to handle and propagate, should be available as a wide variety of genetically defined strains, and should accept a range of vectors. The bacterium Escherichia coli fulfils these requirements and is used in many cloning protocols.Bacteria are mostly used as expression system.
One disadvantage of using an organism such as E. coli as a host for cloning is that it is a prokaryote, and therefore lacks the membrane bound nucleus (and other organelles) found in eukaryotic cells. This means that certain eukaryotic genes may not function in E. coli as they would in their normal environment, which can hamper their isolation by selection mechanisms that depend on gene expression.
Also, if the production of a eukaryotic protein is the desired outcome of a cloning experiment, it may not be easy to ensure that a prokaryotic host produces a fully functional protein.
Eukaryotic cells range from microbes, such as yeast and algae, to cells from complex multicellular organisms, such as ourselves.
In other words, the Genetic engineering is biochemical manipulation of genes by which foreign genes or the genes of donor organisms are inserted into the DNA or (touted recipient organism through the plasmids of bacteria or through bacteriophages.
Historical Account of Genetic Engineering:In nature the transfer of genetic material occurs through conjugation, transformation and transduction.
Recombination is brought about by the following two processes:
(i) Natural recombination process, and
(ii) Artificial recombination process.
The natural recombination occurs during meiosis when similar nucleotide sequences in DNAs of homologous chromosomes come closer, and exchange segments and then rejoin. Thus a new arrangement of genes results from this recombination.
The artificial recombination process involves unrelated organisms. The genetic material of one organism is inserted into a specialized DNA molecule of another organism giving rise to recombinant or rDNA.
The history of modern genetic engineering goes back to mid 1970s when it became possible to cut DNA into pieces and transfer a particular piece of DNA containing specific information from donor organism to the DNA of the recipient organism.
And year-wise landmarks of the genetic engineering are given here as follows:
1970 Smith and Nathans discovered a new class of enzymes, the restriction enzymes that act as chemical scissors and cut a DNA molecule into smaller fragments.1972 Berg and others combined DNAs from two viruses to produce what is called recombinant DNA (rDNA).
1973 Cohen and Boyer inserted recombinant DNAs into host bacteria that reproduced or cloned foreign DNAs. With this, the age of genetic engineering begins.
1977 Genetech, one of the first genetic engineering companies starts the biosynthesis of important drugs by rDNA Technology.
1977 Sanger and Gilbert independently discovered techniques for rapid sequencing of nucleotides in DNA molecules.
1982 Human insulin produced by rDNA is marketted under the trade name ‘Humulin’.
1983 Tracy Moreno, a ten year old girl born with growth hormone deficiency, grows 5 inches in one year of treatment with engineered hormones. Human growth hormone genes are inserted into mouse embryo producing a giant mouse. For the first time a human gene functions in another animal.
1997 Scientists remove the DNA containing nucleus from a female’s egg and replace it with a nucleus from a different animal of the same species. The scientists then place that egg into the uterus of a third animal.
The result, first demonstrated by the birth of a cloned sheep named Dolly, is the birth of an animal that is nearly genetically identical to the animal from which the nucleus was obtained. Such an animal is genetically unrelated to the surrogate mother.
Such experiments were also done to produce the clone of human beings but....
In February, 2003 the Human Cloning Prohibition Act was passed which made it a crime for any one, public or private, to conduct somatic cell nuclear transplantation on fertilized or unfertilized human egg cell for reproductive or therapeutic purposes. The penalty for engaging in such research is US $ 1 million fine and a jail term for 10 years.
 |
| Recombinant DNA technology |
Steps of Genetic Engineering: The gene cloning or recombinant DNA formation requires the following
1. DNA fragments to be cloned or target gene sequence of desired type.
2. Restriction endonucleases for cutting DNA molecule into fragments.
3. Cloning vector.
4. DNA ligase enzyme for splicing DNA segments.
5.Expression system
The major steps involved in the genetic engineering are as follows
1. To break open the living cells:Several methods are available to break open the living cells. One of the popular methods involves mechanical shearing, the cells in a blender and then treating them with a detergents.
2. Isolation and identification of desired genes or DNA sequence:
The genetic information is stored in DNA. Since the DNA molecules are much longer than all other molecules found in the cells, it has become possible to develop technique of purifying DNA. DNA molecules are spooled on a glass rod. The glass rod bearing the DNA molecules is then lifted out from the mixture of broken cells.
3. Cutting of the DNA molecules into segments containing specific genes from the rest of DNA:
DNA is cut into gene size segments with the help of molecular scissors,called restriction endonucleases.
The restriction enzymes, which cut DNA at defined sites, represent one of the most important groups of enzymes for the manipulation of DNA. These enzymes are found in bacterial cells, where they function as part of a protective mechanism called the restriction-modification system. In this system the restriction enzyme hydrolyses any exogenous DNA that appears in the cell. To prevent the enzyme acting on the host cell DNA, the modification enzyme of the system (a methylase) modifies the host DNA by methylation of particular bases in the restriction enzyme’s recognition sequence, which prevents the enzyme from cutting the DNA.
Restriction enzymes are of three types (I, II, or III). Most of the enzymes commonly used today are type II enzymes.These enzymes are nucleases, and as they cut at an internal position in a DNA strand (as opposed to beginning degradation at one end) they are known as endonucleases. Thus, the correct designation of such enzymes is that they are type II restriction endonucleases, although they are often simply called restriction enzymes. In essence they may be thought of as molecular scissors.
Restriction enzyme nomenclature
Restriction enzyme nomenclature is based on a number of conventions. The generic and specific names of the organism in which the enzyme is found are used to provide the first part of the designation, which comprises the first letter of the generic name and the first two letters of the specific name.Thus, an enzyme from a strain of Escherichia coli is termed Eco, one from Bacillus amyloliquefaciens is Bam, and so on.
Use of restriction endonucleases
Restriction enzymes are very simple to use -- an appropriate amount of enzyme is added to the target DNA in a buffer solution, and the reaction is incubated at the optimum temperature (usually 37◦C) for a suitable length of time. Enzyme activity is expressed in units, with one unit being the amount of enzyme that will cleave one microgram of DNA in one hour at 37◦C.
Type of restriction endonucleases on the base of activityThere are two groups of restriction enzymes. Some enzymes recognise a specific nucleotide pair sequence and then cut the DNA at random i.e., at non-specific site away from the recognition site.
The other group of enzymes cleave the DNA at specific site. All restriction enzymes are sequence specific and thus the number of cuts they make in a DNA molecule depends upon the number of times, the specific sequences are repeated in DNA molecules. Different endonucleases found in different organisms recognise different nucleotide sequences and they cut DNA at different cleavage sites.
Due to the activities of restriction enzymes following types of DNA fragments are produced:
(1) blunt ends (sometimes known as flush-ended fragments), (2) fragments with protruding 3 ends, and (3) fragments with protruding 5 ends.
Enzymes such as EcoRI generate DNA fragments with cohesive or ‘sticky’ ends, as the protruding sequences can base pair with complementary sequences generated by the same enzyme. Thus, by cutting two different DNA samples with the same enzyme and mixing the fragments together, recombinant DNA can be produced. This is one of the most useful applications of restriction enzymes and is a vital part of many manipulations in genetic engineering.
3.Use of a vector
The biology of gene cloning is concerned with the selection and use of a suitable carrier molecule or vector, and a living system or host in which the vector can be propagated.
The means by which recombinant DNA is introduced into a host cell is called vector.
There are certain features that vectors must posess. Ideally they should be fairly small DNA molecules, to facilitate isolation and handling. There must be an origin of replication, so that their DNA can be copied and thus maintained in the cell population as the host organism grows and divides. It is desirable to have some sort of selectable marker that will enable the vector to be detected, and the vector must also have at least one unique restriction endonuclease recognition site to enable DNA to be inserted during the production of recombinants. Plasmids have these features and are extensively used as vectors in cloning experiment.
Plasmids:Many types of plasmids are found in nature, in bacteria and some yeasts. They are circular extrachromosomal DNA molecules carrying genes for antibiotic resistance and fertility in bacteria.
Plasmids can be classified into two groups, conjugative and non-conjugative plasmids. Conjugative plasmids can mediate their own transfer between bacteria by the process of conjugation,Non-conjugative plasmids are not selftransmissible but may be mobilised by a conjugation-proficient plasmid if their mobilising region is functional.
DNA of bacterial viruses
The DNA of bacterial virus can also be used as a vector.For example lambda phage.Lambda phage attaches to a host bacterium.The recombinant DNA is released from the virus and enters into the bacterium.The recombinant DNA replicates and many copies of the viruses are formed.Each virus in bacteriophage clone contains a copy of gene of choice.
The bacteriophage-based vectors are more specialised than plasmid vectors.Two types of bacteriophage (lambda and M13) have been extensively developed for cloning purposes
DNA ligase – joining DNA molecules
DNA ligase is an important cellular enzyme, as its function is to repair broken phosphodiester bonds that may occur at random or
as a consequence of DNA replication or recombination. In genetic engineering it is used to seal discontinuities in the sugar--phosphate chains that arise when recombinant DNA is made by joining DNA molecules from different sources. It can therefore be thought of as
molecular glue, which is used to stick pieces of DNA together. This function is crucial to the success of many experiments, and DNA ligase is therefore a key enzyme in genetic engineering.
The enzyme used most often in experiments is T4 DNA ligase, which is purified from E. coli cells infected with bacteriophage T4.
Expression system or Host An ideal host cell should be easy to handle and propagate, should be available as a wide variety of genetically defined strains, and should accept a range of vectors. The bacterium Escherichia coli fulfils these requirements and is used in many cloning protocols.Bacteria are mostly used as expression system.
One disadvantage of using an organism such as E. coli as a host for cloning is that it is a prokaryote, and therefore lacks the membrane bound nucleus (and other organelles) found in eukaryotic cells. This means that certain eukaryotic genes may not function in E. coli as they would in their normal environment, which can hamper their isolation by selection mechanisms that depend on gene expression.
Also, if the production of a eukaryotic protein is the desired outcome of a cloning experiment, it may not be easy to ensure that a prokaryotic host produces a fully functional protein.
Eukaryotic cells range from microbes, such as yeast and algae, to cells from complex multicellular organisms, such as ourselves.
 |
| Genetic engineering |






No comments:
Post a Comment
Thank you for commenting.